Human genetic variation is the natural variation in gene frequencies observed between the genomes of individuals or groups of humans. Variation can be measured at both the individual level (differences between individual people) and at the population level, i.e. differences between populations living in different regions.
In genetics there may be multiple variants of any given gene (polymorphism), these are called alleles. Any individual human has only two copies of any given allele, one inherited from their mother and the other from their father, but many more different versions of the gene may exist locally or within their own family, with offspring sharing 50% of their genes with each parent, and 50% of their genes on average with each sibling (see Coefficient of relationship). In a given population certain alleles may be more abundant than other alleles, leading to variation in the frequencies of alleles between populations, the more geographically distant these populations are from each other, the more differences there are between the populations.
There are at least two reasons why genetic variation is geographically distributed:
- natural selection may confer an adaptive advantage to individuals in a specific environment, for example dark skin pigmentation protects from high levels of ultraviolet radiation, whereas a low level of melanin in the skin may confer an advantage in regions with low levels of UV light. Alleles under selection are likely to occur only in those geographic regions where they confer an advantage.
- The second main cause of geographically distributed genetic variation is due to non-uniform sampling of a population. The main cause is founder effect, this is the effect of a small group of individuals migrating from a larger group and founding a new population, if the migrating population represents only a small subset of the parental population, then it will not be genetically representative of the parental population (sampling error). Small founding populations are also subject to genetic drift, which may further alter allele frequencies. An example of this is the human migration out of Africa, it has been theorised that the migration out of Africa only represented a small fraction of the genetic variation in East Africa, and that this is the cause of the observed lower levels of diversity in all indigenous non-African humans.
Generally, more recent neutral polymorphisms caused by mutation are likely to be relatively geographically localised, while older polymorphisms are more likely to be shared by all human groups. A large majority of the observed genetic variation is nevertheless distributed within any geographic region rather than between regions, though it is usually possible to accurately identify the geographic origins of any individual's ancestors by genetic means.
The study of human geographic variation has both evolutionary significance and medical applications. The study can help scientists understand ancient human population migrations as well as how different human groups are biologically related to one another. From a medical perspective the study of human genetic variation may be important because some disease causing alleles occur at a greater frequency in people from specific geographic regions.
Extent of human variation
"Genetic variation among individual humans occurs on many different scales, ranging from gross alterations in the human karyotype to single nucleotide changes." Nucleotide diversity is based on single mutations (single nucleotide polymorphisms (SNPs)); the nucleotide diversity between humans is about 0.1%, that is one difference per 1000 base pairs. This difference between humans is considered small relative to other large primates, for example it is often cited that although chimpanzees have a restricted geographical range and small population numbers their nucleotide diversity is greater than that of humans, with one difference between individuals per 500 base pairs. This is often taken as evidence for the Recent African origin of our species; it also makes it difficult to claim any great divergence between individuals or groups of humans due to our overall relative homogeneity as a species, both on an individual and a group basis.
More recently a better understanding of the structure of the genome has been gained with the publication of two examples of full sequences of an individual's genome. This represents a new development because the human genome project sequence represented two haploid sequences, both of which were an amalgamation of sequences from many individuals. Recently the diploid sequences of both Craig Venter and James Watson have been published. Analysis of diploid sequences has shown that non-SNP variation accounts for much more human genetic variation than single nucleotide diversity. This non-SNP variation is called copy number variation and results from deletions, inversions, insertions and duplications. It is estimated that approximately 0.4% of the genomes of unrelated people typically differ with respect to copy number. When copy number variation is included, human to human genetic variation is estimated to be at least 0.5% (99.5% similarity). Copy number variations, that result in interindividual differences, are not necessarily completely inherited, but can also arise during development.
Distribution of variation
Data gathered to date suggest that human variation exhibits several distinctive characteristics. First, compared with many other mammalian species, humans are genetically less diverse—a counterintuitive finding, given our large population and worldwide distribution (Li and Sadler 1991; Kaessmann et al. 2001). For example, the chimpanzee subspecies living just in central and western Africa have higher levels of diversity than do humans (Ebersberger et al. 2002; Yu et al. 2003; Fischer et al. 2004).
Two random humans are expected to differ at approximately 1 in 1000 nucleotides, whereas two random chimpanzees differ at 1 in 500 nucleotide pairs. However, with a genome of approximate 3 billion nucleotides, on average two humans differ at approximately 3 million nucleotides. Most of these single nucleotide polymorphisms (SNPs) are neutral, but some are functional and influence the phenotypic differences between humans. It is estimated that about 10 million SNPs exist in human populations, where the rarer SNP allele has a frequency of at least 1% (see International HapMap Project).
The distribution of variants within and among human populations also differs from that of many other species. The details of this distribution are impossible to describe succinctly because of the difficulty of defining a "population," the clinal nature of variation, and heterogeneity across the genome (Long and Kittles 2003). In general, however, 6%–10% of genetic variation occurs between large groups living on different continents, with 5%-6% distributed between localised populations within the same continent, the remaining ~85% of the variation exists within populations. (Lewontin 1972; Jorde et al. 2000a; Hinds et al. 2005). Long and Kittles (2003) point out that this estimate is somewhat misleading, in fact the figure of ~85% of diversity existing within populations is an average for all human populations. The recent African origin theory for humans would predict that in Africa there exists a great deal more diversity than without Africa, and that diversity should decrease the further from Africa a population is sampled. Long and Kittles show that indeed, African populations contain about 100% of human genetic diversity, whereas in populations outside of Africa diversity is much reduced, for example in their population from New Guinea only about 70% of human variation is captured. This distribution of genetic variation differs from the pattern seen in many other mammalian species, for which existing data suggest greater differentiation between groups (Templeton 1998; Kittles and Weiss 2003).
Our history as a species also has left genetic signals in regional populations. For example, in addition to having higher levels of genetic diversity, populations in Africa tend to have lower amounts of linkage disequilibrium than do populations outside Africa, partly because of the larger size of human populations in Africa over the course of human history and partly because the number of modern humans who left Africa to colonize the rest of the world appears to have been relatively low (Gabriel et al. 2002). In contrast, populations that have undergone dramatic size reductions or rapid expansions in the past and populations formed by the mixture of previously separate ancestral groups can have unusually high levels of linkage disequilibrium (Nordborg and Tavare 2002).
In the field of population genetics, it is believed that the distribution of neutral polymorphisms among contemporary humans reflects human demographic history. It is believed that humans passed through a population bottleneck before a rapid expansion coinciding with migrations out of Africa leading to an African-Eurasian divergence around 100,000 years ago (ca. 5,000 generations), followed by a European-Asian divergence about 40,000 years ago (ca. 2,000 generations). Richard G. Klein, Nicholas Wade and Spencer Wells, among others, have postulated that modern humans did not leave Africa and successfully colonize the rest of the world until as recently as 60,000 - 50,000 years B.P., pushing back the dates for subsequent population splits as well.
The rapid expansion of a previously small population has two important effects on the distribution of genetic variation. First, the so-called founder effect occurs when founder populations bring only a subset of the genetic variation from their ancestral population. Second, as founders become more geographically separated, the probability that two individuals from different founder populations will mate becomes smaller. The effect of this assortative mating is to reduce gene flow between geographical groups, and to increase the genetic distance between groups. The expansion of humans from Africa affected the distribution of genetic variation in two other ways. First, smaller (founder) populations experience greater genetic drift because of increased fluctuations in neutral polymorphisms. Second, new polymorphisms that arose in one group were less likely to be transmitted to other groups as gene flow was restricted.
Many other geographic, climatic, and historical factors have contributed to the patterns of human genetic variation seen in the world today. For example, population processes associated with colonization, periods of geographic isolation, socially reinforced endogamy, and natural selection all have affected allele frequencies in certain populations (Jorde et al. 2000b; Bamshad and Wooding 2003). In general, however, the recency of our common ancestry and continual gene flow among human groups have limited genetic differentiation in our species.
Molecular lineages, Y chromosomes and mitochondrial DNA
Mitochondria are small organelles that lie in the cytoplasm of eucaryotic cells, such as those of humans. Their primary purpose is to provide energy to the cell. Mitochondria are thought to be the vestigial remains of symbiotic bacteria that were once free living. One indication that mitochondria were once free living is that they contain a relatively small circular segment of DNA, called mitochondrial DNA (mtDNA). The overwhelming majority of a human's DNA is contained in chromosomes in the nucleus of the cell, but mtDNA is an exception. An individual inherits their cytoplasm and the organelles it contains exclusively from their mother, as these are derived from the ovum (egg cell), sperm only carry chromosomal DNA due to the necessity of maintaining motility. When a mutation arises in mtDNA molecule, the mutation is therefore passed in a direct female line of descent. These mutations are derived from copying mistakes, when the DNA is copied it is possible that a single mistake occurs in the DNA sequence, these single mistakes are called single nucleotide polymorphisms (SNPs).
Human Y chromosomes are male-specific sex chromosomes; any human that possesses a Y chromosome will be morphologically male. Y chromosomes are therefore passed from father to son; although Y chromosomes are situated in the cell nucleus, they only recombine with the X chromosome at the ends of the Y chromosome; the vast majority of the Y chromosome (95%) does not recombine. Therefore, as with mtDNA, when mutations (SNPs) arise in the Y chromosome, they are passed on directly from father to son in a direct male line of descent.
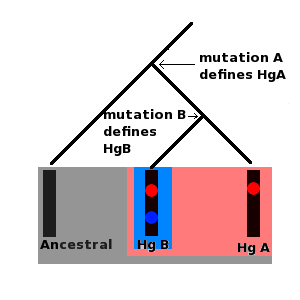 Ancestral Haplogroup Haplogroup A (Hg A) Haplogroup B (Hg B) All of these molecules are part of the ancestral haplogroup, but at some point in the past a mutation occurred in the ancestral molecule, mutation A, which produced a new lineage; this is haplogroup A and is defined by mutation A. At some more recent point in the past, a new mutation, mutation B, occurred in a person carrying haplogroup A; mutation B defined haplogroup B. Haplogroup B is a subgroup, or subclade of haplogroup A; both haplogrups A and B are subclades of the ancestral haplogroup.
Ancestral Haplogroup Haplogroup A (Hg A) Haplogroup B (Hg B) All of these molecules are part of the ancestral haplogroup, but at some point in the past a mutation occurred in the ancestral molecule, mutation A, which produced a new lineage; this is haplogroup A and is defined by mutation A. At some more recent point in the past, a new mutation, mutation B, occurred in a person carrying haplogroup A; mutation B defined haplogroup B. Haplogroup B is a subgroup, or subclade of haplogroup A; both haplogrups A and B are subclades of the ancestral haplogroup. The Y chromosome and mtDNA therefore share certain properties. Other chromosomes, autosomes and X chromosomes in women, share their genetic material (called crossing over leading to recombination) during meiosis (a special type of cell division that occurs for the purposes of sexual reproduction). Effectively this means that the genetic material from these chromosomes gets mixed up in every generation, and so any new mutations are passed down randomly from parents to offspring. The special feature that both Y chromosomes and mtDNA display is that mutations can accrue along a certain segment of both molecules and these mutations remain fixed in place on the DNA. Furthermore the historical sequence of these mutations can also be inferred. For example, if a set of ten Y chromosomes (derived from ten different men) contains a mutation, A, but only five of these chromosomes contain a second mutation, B, it must be the case that mutation B occurred after mutation A. Furthermore all ten men who carry the chromosome with mutation A are the direct male line descendants of the same man who was the first person to carry this mutation. The first man to carry mutation B was also a direct male line descendant of this man, but is also the direct male line ancestor of all men carrying mutation B. Series of mutations such as this form molecular lineages. Furthermore each mutation defines a set of specific Y chromosomes called a haplogroup. All men carrying mutation A form a single haplogroup, all men carrying mutation B are part of this haplogroup, but mutation B also defines a more recent haplogroup (which is a subgroup or subclade) of its own which men carrying only mutation A do not belong to. Both mtDNA and Y chromosomes are grouped into lineages and haplogroups; these are often presented as tree like diagrams.
Groundbreaking work by molecular biologists such as Cann et al. (1987) on mtDNA produced three interesting observations relevant to race and human evolution.
Firstly, by estimating the rate at which mutations occur in mtDNA Cann et al. were able to estimate the age of the common ancestral mtDNA type: "the common ancestral mtDNA (type a) links mtDNA types that have diverged by an average of nearly 0.57%. Assuming a rate of 2%-4% per million years, this implies that the common ancestor of all surviving mtDNA types existed 140,000-290,000 years ago." This observation is robust, and this common direct female line ancestor (or mitochondrial most recent common ancestor (mtMRCA)) of all extant humans has become known as mitochondrial eve. The observation that the mtMRCA is the direct matrilineal ancestor of all living humans should not be interpreted as meaning that either she was the first anatomically modern human, nor that there were no other female humans living concurrently with her. A more reasonable explanation is that other women who lived at the same time as mtMRCA did indeed reproduce and pass their genes down to living humans, but that their mitochondrial lineages have been lost over time, probably due to random events such as producing only male children. It is impossible to know to what extent these non-extant lineages have been lost or how much they differed from the mtDNA of our mtMRCA. Cann et al.
Secondly, Cann et al. postulate that their work supports an African origin for modern human mtDNA: "We infer from the tree of minimum length... that Africa is a likely source of the human mitochondrial gene pool. This inference comes from the observation that one of the two primary branches leads exclusively to African mtDNAs... while the second primary branch also leads to African mtDNAs... By postulating that the common ancestral mtDNA (type a in Fig. 3) was African, we minimize the number of intercontinental migrations needed to account for the geographic distribution of mtDNA types."
Thirdly, the study shows that mtDNA types (haplogroups) do not cluster by geography, ethnicity or race, implying multiple female lineages were involved in founding modern human populations, with many closely related lineages spread geographically and many populations containing distantly related lineages: "The second implication of the tree (Fig. 3)—that each non-African population has multiple origins—can be illustrated most simply with the New Guineans. Take, as an example, mtDNA type 49, a lineage whose nearest relative is not in New Guinea, but in Asia (type 50). Asian lineage 50 is closer genealogically to this New Guinea lineage than to other Asian mtDNA lineages. Six other lineages lead exclusively to New Guinean mtDNAs, each originating at a different place in the tree (types 12, 13, 26-29, 65, 95 and 127-134 in Fig. 3). This small region of New Guinea (mainly the Eastern Highlands Province) thus seems to have been colonised by at least seven maternal lineages (Tables 2 and 3). In the same way, we calculate the minimum numbers of female lineages that colonised Australia, Asia and Europe (Tables 2 and 3). Each estimate is based on the number of region-specific clusters in the tree (Fig. 3, Tables 2 and 3). These numbers, ranging from 15 to 36 (Tables 2 and 3), will probably rise as more types of human mtDNA are discovered."
The Y chromosome is much larger than mtDNA, and is relatively homogeneous; therefore it has taken much longer to find distinct lineages and to analyse them. Conversely, because the Y chromosome is so large by comparison, it can hold a great deal more genetic information. With regard to the three observations made by Cann et al. concerning mtDNA, Y chromosome studies show similar patterns. The estimate for the age of the ancestral Y chromosome for all extant Y chromosomes is given at about 70,000 years ago and is also placed in Africa, this individual is sometimes referred to as Y chromosome Adam. The difference in dates between Y chromosome Adam and mitochondrial Eve is usually attributed to a higher extinction rate for Y chromosomes due to greater differential reproductive success between individual men, which means that a small number of very successful men may produce a great many children, while a larger number of less successful men will produce far less children. Keita et al. (2004) say, with reference to Y chromosome and mtDNA and concepts of race:
Y-chromosome and mitochondrial DNA genealogies are especially interesting because they demonstrate the lack of concordance of lineages with morphology and facilitate a phylogenetic analysis. Individuals with the same morphology do not necessarily cluster with each other by lineage, and a given lineage does not include only individuals with the same trait complex (or 'racial type'). Y-chromosome DNA from Africa alone suffices to make this point. Africa contains populations whose members have a range of external phenotypes. This variation has usually been described in terms of 'race' (Caucasoids, Pygmoids, Congoids, Khoisanoids). But the Y-chromosome clade defined by the PN2 transition (PN2/M35, PN2/M2) [see haplogroup E3b and Haplogroup E3a] shatters the boundaries of phenotypically defined races and true breeding populations across a great geographical expanse. African peoples with a range of skin colors, hair forms and physiognomies have substantial percentages of males whose Y chromosomes form closely related clades with each other, but not with others who are phenotypically similar. The individuals in the morphologically or geographically defined 'races' are not characterized by 'private' distinct lineages restricted to each of them.
How much are genes shared? Clustering analyses and what they tell us
Genetic data can be used to infer population structure and assign individuals to groups that often correspond with their self-identified geographical ancestry. Recently, Lynn Jorde and Steven Wooding argued that "Analysis of many loci now yields reasonably accurate estimates of genetic similarity among individuals, rather than populations. Clustering of individuals is correlated with geographic origin or ancestry."
In 2003 A. W. F. Edwards wrote a paper called Lewontin's Fallacy, rebuking the argument that because most of the variation is within-group, therefore classification of humans is not possible. He claimed that this conclusion ignores the fact that most of the information that distinguishes populations is hidden in the correlation structure of the data and not simply in the variation of the individual factors. Edwards concludes that "It is not true that 'racial classification is ... of virtually no genetic or taxonomic significance' or that 'you can't predict someone’s race by their genes'."Likewise Neil Risch of Stanford University has proposed that self-identified race/ethnic group could be a valid means of categorization in the USA for public health and policy considerations. While a 2002 paper by Noah Rosenberg's group makes a similar claim "The structure of human populations is relevant in various epidemiological contexts. As a result of variation in frequencies of both genetic and nongenetic risk factors, rates of disease and of such phenotypes as adverse drug response vary across populations. Further, information about a patient’s population of origin might provide health care practitioners with information about risk when direct causes of disease are unknown."
Researchers such as Neil Risch and Noah Rosenberg have argued that a person's biological and cultural background may have important implications for medical treatment decisions, for example an opinion paper by Neil Risch's group in 2002 states:
Both for genetic and non-genetic reasons, we believe that racial and ethnic groups should not be assumed to be equivalent, either in terms of disease risk or drug response.....Whether African Americans, Hispanics, Native Americans, Pacific Islanders or Asians respond equally to a particular drug is an empirical question that can only be addressed by studying these groups individually.
While another 2002 paper by Noah Rosenberg's group makes a similar claim
The structure of human populations is relevant in various epidemiological contexts. As a result of variation in frequencies of both genetic and nongenetic risk factors, rates of disease and of such phenotypes as adverse drug response vary across populations. Further, information about a patient’s population of origin might provide health care practitioners with information about risk when direct causes of disease are unknown.
This work used samples from the Human Genome Diversity Project (HGDP), a project that has collected samples from individuals from 52 ethnic groups from various locations around the world. The HGDP has itself been criticised for collecting samples on an "ethnic group" basis, on the grounds that ethnic groups represent constructed categories rather than categories which are solely natural or biological. The molecular anthropologist Jonathan Marks states:
As any anthropologist knows, ethnic groups are categories of human invention, not given by nature. Their boundaries are porous, their existence historically ephemeral. There are the French, but no more Franks; there are the English, but no Saxons; and Navajos, but no Anasazi...we cannot really know the nature of the actual relationship of the modern group to the ancient one...The worst mistake you can make in human biology is to confuse constructed categories with natural ones. And to overload a big project with cultural categories as the overall sampling strategy would be a serious problem
In the same issue of Science that published the Rosenberg data, Mary-Claire King and Arno G. Motulsky give a similar warning regarding the HGDP data:
The identification of clusters corresponding to the major geographic regions may depend on the sampling of individuals from well-defined, relatively homogeneous populations. If individuals were sampled from a worldwide 'grid' (or a worldwide grid weighted by population density), the clusters might be much less precisely defined. Does the correspondence of worldwide genetic clusters and major geographic regions suggest borders around genetic clusters analogous to the physical borders—oceans, mountain ranges, and deserts—separating geographic regions? No. Both the results of Rosenberg and colleagues and those of previous studies indicate that unlike separations between geographic regions, differences in allele frequencies are gradual.
Another study by Neil Risch in 2005 used 326 microsatellite markers and self-identified race/ethnic group (SIRE), white (European American), African-American (black), Asian and Hispanic (individuals involved in the study had to choose from one of these categories), to representing discrete "populations", and showed distinct and non-overlapping clustering of the white, African-American and Asian samples. The results were claimed to confirm the integrity of self-described ancestry: "We have shown a nearly perfect correspondence between genetic cluster and SIRE for major ethnic groups living in the United States, with a discrepancy rate of only 0.14%." But also warned that: "This observation does not eliminate the potential for confounding in these populations. First, there may be subgroups within the larger population group that are too small to detect by cluster analysis. Second, there may not be discrete subgrouping but continuous ancestral variation that could lead to stratification bias. For example, African Americans have a continuous range of European ancestry that would not be detected by cluster analysis but could strongly confound genetic case-control studies. (Tang, 2005)
Studies such as those by Risch and Rosenberg use a computer program called STRUCTURE to find human populations (gene clusters). It is a statistical program that works by placing individuals into one of two clusters based on their overall genetic similarity, many possible pairs of clusters are tested per individual to generate multiple clusters. These populations are based on multiple genetic markers that are often shared between different human populations even over large geographic ranges. The notion of a genetic cluster is that people within the cluster share on average similar allele frequencies to each other than to those in other clusters. (Edwards, 2003 but see also infobox "Multi Locus Allele Clusters") In a test of idealised populations, the computer programme STRUCTURE was found to consistently under-estimate the numbers of populations in the data set when high migration rates between populations and slow mutation rates (such as single nucleotide polymorphisms) were considered.
Nevertheless the Rosenberg et al. (2002) paper shows that individuals can be assigned to specific clusters to a high degree of accuracy. One of the underlying questions regarding the distribution of human genetic diversity is related to the degree to which genes are shared between the observed clusters. It has been observed repeatedly that the majority of variation observed in the global human population is found within populations. This variation is usually calculated using Sewall Wright's Fixation index (FST), which is an estimate of between to within group variation. The degree of human genetic variation is a little different depending upon the gene type studied, but in general it is common to claim that ~85% of genetic variation is found within groups, ~6-10% between groups within the same continent and ~6-10% is found between continental groups. For example The Human Genome Project states "two random individuals from any one group are almost as different [genetically] as any two random individuals from the entire world." On the other hand Edwards (2003) claims in his essay "Lewontin's Fallacy" that: "It is not true, as Nature claimed, that 'two random individuals from any one group are almost as different as any two random individuals from the entire world'" and Risch et al. (2002) state "Two Caucasians are more similar to each other genetically than a Caucasian and an Asian." It should be noted that these statements are not the same. Risch et al. simply state that two indigenous individuals from the same geographical region are more similar to each other than either is to an indigenous individual from a different geographical region, a claim few would argue with. Jorde et al put it like this:
The picture that begins to emerge from this and other analyses of human genetic variation is that variation tends to be geographically structured, such that most individuals from the same geographic region will be more similar to one another than to individuals from a distant region.
Whereas Edwards claims that it is not true that the differences between individuals from different geographical regions represent only a small proportion of the variation within the human population (he claims that within group differences between individuals are not almost as large as between group differences). Bamshad et al. (2004) used the data from Rosenberg et al. (2002) to investigate the extent of genetic differences between individuals within continental groups relative to genetic differences between individuals between continental groups. They found that though these individuals could be classified very accurately to continental clusters, there was a significant degree of genetic overlap on the individual level, to the extent that, using 377 loci, individual Europeans were about 38% of the time more genetically similar to East Asians than to other Europeans.
The results obtained by clustering analyses are dependent on several criteria:
- The clusters produced are relative clusters and not absolute clusters, each cluster is the product of comparisons between sets of data derived for the study, results are therefore highly influenced by sampling strategies. (Edwards, 2003)
- The geographic distribution of the populations sampled, because human genetic diversity is marked by isolation by distance, populations from geographically distant regions will form much more discrete clusters than those from geographically close regions. (Kittles and Weiss, 2003)
- The number of genes used. The more genes used in a study the greater the resolution produced and therefore the greater number of clusters that will be identified. (Tang, 2005)
Additionally two studies of European population clusters have been produced. Seldin et al. (2006) identified three European clusters using 5,700 genome-wide polymorphisms. Bauchet et al. (2007) used 10,000 polymorphisms to identify five distinct clusters in the European population, consisting of a south-eastern European cluster (including samples from southern Italians, Armenian, Ashkenazi Jewish and Greek "populations"); a northern-European Cluster (including samples from German, eastern English, Polish and western Irish "populations"); a Basque cluster (including samples from Basque "populations"); a Finnish cluster (including samples from Finnish "populations") and a Spanish cluster (including samples from Spanish "populations"). Most "populations" contained individuals from clusters other than the dominant cluster for that population, there were also individuals with membership of several clusters. The results of this study are presented on a map of Europe. (Bauchet, 2007) The existence of allelic clines and the observation that the bulk of human variation is continuously distributed, has led some scientists to conclude that any categorization schema attempting to partition that variation meaningfully will necessarily create artificial truncations. (Kittles & Weiss 2003). It is for this reason, Reanne Frank argues, that attempts to allocate individuals into ancestry groupings based on genetic information have yielded varying results that are highly dependent on methodological design. Serre and Pääbo (2004) make a similar claim:
The absence of strong continental clustering in the human gene pool is of practical importance. It has recently been claimed that “the greatest genetic structure that exists in the human population occurs at the racial level” (Risch et al. 2002). Our results show that this is not the case, and we see no reason to assume that “races” represent any units of relevance for understanding human genetic history.
In a response to Serre and Pääbo (2004), Rosenberg et al. (2005) make three relevant observations. Firstly they maintain that their clustering analysis is robust. Secondly they agree with Serre and Pääbo that membership of multiple clusters can be interpreted as evidence for clinality (isolation by distance), though they also comment that this may also be due to admixture between neighbouring groups (small island model). Thirdly they comment that evidence of clusterdness is not evidence for any concepts of "biological race".
Serre and Pääbo argue that human genetic diversity consists of clines of variation in allele frequencies. We agree and had commented on this issue in our original paper: “In several populations, individuals had partial membership in multiple clusters, with similar membership coefficients for most individuals. These populations might reflect continuous gradations across regions or admixture of neighboring groups.” (Rosenberg, 2002) At the same time, we find that human genetic diversity consists not only of clines, but also of clusters, which STRUCTURE observes to be repeatable and robust....Our evidence for clustering should not be taken as evidence of our support of any particular concept of “biological race.” In general, representations of human genetic diversity are evaluated based on their ability to facilitate further research into such topics as human evolutionary history and the identification of medically important genotypes that vary in frequency across populations. Both clines and clusters are among the constructs that meet this standard of usefulness: for example, clines of allele frequency variation have proven important for inference about the genetic history of Europe, and clusters have been shown to be valuable for avoidance of the false positive associations that result from population structure in genetic association studies. The arguments about the existence or nonexistence of “biological races” in the absence of a specific context are largely orthogonal to the question of scientific utility, and they should not obscure the fact that, ultimately, the primary goals for studies of genetic variation in humans are to make inferences about human evolutionary history, human biology, and the genetic causes of disease.
Similarly Witherspoon et al. (2007) have shown that while it is possible to classify people into genetic clusters this does not resolve the observation that any two individuals from different populations are often genetically more similar to each other than to two individuals from the same population:
Discussions of genetic differences between major human populations have long been dominated by two facts: (a) Such differences account for only a small fraction of variance in allele frequencies, but nonetheless (b) multilocus statistics assign most individuals to the correct population. This is widely understood to reflect the increased discriminatory power of multilocus statistics. Yet Bamshad et al. (2004) showed, using multilocus statistics and nearly 400 polymorphic loci, that (c) pairs of individuals from different populations are often more similar than pairs from the same population. If multilocus statistics are so powerful, then how are we to understand this finding?
All three of the claims listed above appear in disputes over the significance of human population variation and "race"...The Human Genome Project (2001, p. 812) states that "two random individuals from any one group are almost as different [genetically] as any two random individuals from the entire world."
Risch et al. (2002) state that "two Caucasians are more similar to each other genetically than a Caucasian and an Asian", but Bamshad et al (2004) used the same data set as Rosenberg et al. (2002) to show that Europeans are more similar to Asians 38% of the time than they are to other Europeans when only 377 microsatellite markers are analysed.

Bottom: Distribution of genetic variation if isolation by distance is considered; all variation is gradual over the extent of the landmass. This pattern is "clinal".
In agreement with the observation of Bamshad et al. (2004), Witherspoon et al. (2007) have shown that many more than 326 or 377 microsatellite loci are required in order to show that individuals are always more similar to individuals in their own population group than to individuals in different population groups, even for three distinct populations.
In 2007 Witherspoon et al. sought to investigate these apparently contradictory observations. In their paper Genetic similarities within and between human populations they expand upon the observation of Bamshad et al. (2004). They show that the observed clustering of human populations into relatively discrete groups is a product of using what they call "population trait values". This means that each individual is compared to the "typical" trait for several populations, and assigned to a population based on the individual's overall similarity to one of the populations as a whole: "population membership is treated as an additive quantitative genetic trait controlled by many loci of equal effect, and individuals are divided into populations on the basis of their trait values." They therefore claim that clustering analyses cannot necessarily be used to make inferences regarding the similarity or dissimilarity of individuals between or within clusters, but only for similarities or dissimilarities of individuals to the "trait values" of any given cluster. The paper measures the rate of misclassification using these "trait values" and calls this the "population trait value misclassification rate" (CT). The paper investigates the similarities between individuals by use of what they term the "dissimilarity fraction" (ω): "the probability that a pair of individuals randomly chosen from different populations is genetically more similar than an independent pair chosen from any single population." Witherspoon et al. show that two individuals can be more genetically similar to each other than to the typical genetic type of their own respective populations, and yet be correctly assigned to their respective populations. An important observation is that the likelihood that two individuals from different populations will be more similar to each other genetically than two individuals from the same population depends on several criteria, most importantly the number of genes studied and the distinctiveness of the populations under investigation.
Given 10 loci, three distinct populations, and the full spectrum of polymorphisms, the answer is ω ~ 0.3, or nearly one-third of the time. With 100 loci, the answer is ~20% of the time and even using 1000 loci, ω ~ 10%. However, if genetic similarity is measured over many thousands of loci, the answer becomes never when individuals are sampled from geographically separated populations.
By geographically separated populations, they mean sampling of people only from distant geographical regions while omitting intermediate regions, in this case Europe, sub-Saharan Africa, and East Asian. They continue:
On the other hand, if the entire world population were analyzed, the inclusion of many closely related and admixed populations would increase ω... In a similar vein, Romualdi et al. (2002) and Serre and Paabo (2004) have suggested that highly accurate classification of individuals from continuously sampled (and therefore closely related) populations may be impossible.... Classification methods typically make use of aggregate properties of populations, not just properties of individuals or even of pairs of individuals... The Structure classification algorithm (Pritchard et al. 2000) also relies on aggregate properties of populations, such as Hardy–Weinberg and linkage equilibrium. In contrast, the pairwise distances used to compute ω make no use of population-level information and are strongly affected by the high level of within-groups variation typical of human populations. This accounts for the difference in behavior between ω and the classification results.
Witherspoon et al. also add:
given enough genetic data, individuals can be correctly assigned to their populations of origin is compatible with the observation that most human genetic variation is found within populations, not between them. It is also compatible with our finding that, even when the most distinct populations are considered and hundreds of loci are used, individuals are frequently more similar to members of other populations than to members of their own population.
Substructure in the human population
New data on human genetic variation has reignited the debate surrounding race. Most of the controversy surrounds the question of how to interpret this new data, and whether conclusions based on existing data are sound. A large majority of researchers endorse the view that continental groups do not constitute different subspecies. However, other researchers still debate whether evolutionary lineages should rightly be called "races". These questions are particularly pressing for biomedicine, where self-described race is often used as an indicator of ancestry (see race in biomedicine below).
Although the genetic differences among human groups are relatively small, these differences in certain genes such as duffy, ABCC11, SLC24A5, called ancestry-informative markers (AIMs) nevertheless can be used to reliably situate many individuals within broad, geographically based groupings or self-identified race. For example, computer analyses of hundreds of polymorphic loci sampled in globally distributed populations have revealed the existence of genetic clustering that roughly is associated with groups that historically have occupied large continental and subcontinental regions (Rosenberg et al. 2002; Bamshad et al. 2003).
Some commentators have argued that these patterns of variation provide a biological justification for the use of traditional racial categories. They argue that the continental clusterings correspond roughly with the division of human beings into sub-Saharan Africans; Europeans, Western Asians, Southern Asians and Northern Africans; Eastern Asians, Southeast Asians, Polynesians and Native Americans; and other inhabitants of Oceania (Melanesians, Micronesians & Australian Aborigines) (Risch et al. 2002). Other observers disagree, saying that the same data undercut traditional notions of racial groups (King and Motulsky 2002; Calafell 2003; Tishkoff and Kidd 2004). They point out, for example, that major populations considered races or subgroups within races do not necessarily form their own clusters. Thus, samples taken from India and Pakistan affiliate with Europeans or eastern Asians rather than separating into a distinct cluster.
Furthermore, because human genetic variation is clinal, many individuals affiliate with two or more continental groups. Thus, the genetically based "biogeographical ancestry" assigned to any given person generally will be broadly distributed and will be accompanied by sizable uncertainties (Pfaff et al. 2004).
In many parts of the world, groups have mixed in such a way that many individuals have relatively recent ancestors from widely separated regions. Although genetic analyses of large numbers of loci can produce estimates of the percentage of a person's ancestors coming from various continental populations (Shriver et al. 2003; Bamshad et al. 2004), these estimates may assume a false distinctiveness of the parental populations, since human groups have exchanged mates from local to continental scales throughout history (Cavalli-Sforza et al. 1994; Hoerder 2002). Even with large numbers of markers, information for estimating admixture proportions of individuals or groups is limited, and estimates typically will have wide confidence intervals or CIs (Pfaff et al. 2004).
Epigenetics
Epigenetics is another type of genetic variation. "This type of variation arises from chemical tags that attach to DNA and affect how it gets read. The chemical tags, called epigenetic markings, act as switches that control how genes can be read."
Variation in phenotyp
The distribution of many physical traits resembles the distribution of genetic variation within and between human populations (American Association of Physical Anthropologists 1996; Keita and Kittles 1997). For example, ~90% of the variation in human head shapes occurs within continental groups, and ~10% separates groups, with a greater variability of head shape among individuals with recent African ancestors (Relethford 2002).
Variation in a trait under selection, skin colour
A prominent exception to the common distribution of physical characteristics within and among groups is skin color. Approximately 10% of the variance in skin color occurs within groups, and ~90% occurs between groups (Relethford 2002). This distribution of skin color and its geographic patterning — with people whose ancestors lived predominantly near the equator having darker skin than those with ancestors who lived predominantly in higher latitudes — indicate that this attribute has been under strong selective pressure. Darker skin appears to be strongly selected for in equatorial regions to prevent sunburn, skin cancer, the photolysis of folate, and damage to sweat glands (Sturm et al. 2001; Rees 2003). A leading hypothesis for the selection of lighter skin in higher latitudes is that it enables the body to form greater amounts of vitamin D, which helps prevent rickets (Jablonski 2004). Evidence for this includes the finding that a substantial portion of the differences of skin color between Europeans and Africans resides in a single gene, SLC24A5 the threonine-111 allele of which was found in 98.7 to 100% among several European samples, while the alanine-111 form was found in 93 to 100% of samples of Africans, East Asians and Indigenous Americans (Lamason et al. 2005). However, the vitamin D hypothesis is not universally accepted (Aoki 2002), and lighter skin in high latitudes may correspond simply to an absence of selection for dark skin (Harding et al. 2000). Melanin which serves as the pigment, is located in the epidermis of the skin, and is based on hereditary gene expression.
Because skin color has been under strong selective pressure, similar skin colors can result from convergent adaptation rather than from genetic relatedness. Sub-Saharan Africans, populations from southern India, and Indigenous Australians have similar skin pigmentation, but genetically they are no more similar than are other widely separated groups. Furthermore, in some parts of the world in which people from different regions have mixed extensively, the connection between skin color and ancestry has been substantially weakened (Parra et al. 2004). In Brazil, for example, skin color is not closely associated with the percentage of recent African ancestors a person has, as estimated from an analysis of genetic variants differing in frequency among continent groups (Parra et al. 2003).
Considerable speculation has surrounded the possible adaptive value of other physical features characteristic of groups, such as the constellation of facial features observed in many eastern and northeastern Asians (Guthrie 1996). However, any given physical characteristic generally is found in multiple groups (Lahr 1996), and demonstrating that environmental selective pressures shaped specific physical features will be difficult, since such features may have resulted from sexual selection for individuals with certain appearances or from genetic drift (Roseman 2004).














No comments:
Post a Comment