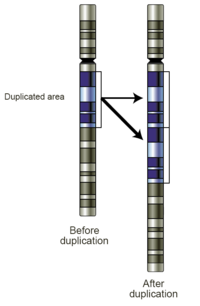
Gene duplication (or chromosomal duplication or gene amplification) is any duplication of a region of DNA that contains a gene; it may occur as an error in homologous recombination, a retrotransposition event, or duplication of an entire chromosome. The second copy of the gene is often free from selective pressure — that is, mutations of it have no deleterious effects to its host organism. Thus it mutates faster than a functional single-copy gene, over generations of organisms.
Gene duplication as an evolutionary event
Gene duplication is believed to play a major role in evolution; this stance has been held by members of the scientific community for over 100 years. Susumu Ohno was one of the most famous developers of this theory in his classic book Evolution by gene duplication (1970). Ohno argued that gene duplication is the most important evolutionary force since the emergence of the universal common ancestor. Major genome duplication events are not uncommon. It is believed that the entire yeast genome underwent duplication about 100 million years ago. Plants are the most prolific genome duplicators. For example, wheat is hexaploid (a kind of polyploid), meaning that it has six copies of its genome.
The two genes that exist after a gene duplication event are called paralogs and usually code for proteins with a different function and/or structure. By contrast, orthologous genes are ones which code for proteins with similar functions but exist in different species, and are created from a speciation event. (See Homology of sequences in genetics).
It is important (but often difficult) to differentiate between paralogs and orthologs in biological research. Experiments on human gene function can often be carried out on other species if a homolog to a human gene can be found in the genome of that species, but only if the homolog is orthologous. If they are paralogs and resulted from a gene duplication event, their functions are likely to be too different.
The paralogous segments can be repeat sequences with more than 90% sequence similarity. In such cases, they are known as low copy repeats (LCRs) though they are not highly repetitive sequences. They are mostly found in pericentronomic, subtelomeric and interstitial regions of a chromosome. The LCRs, due to their size (>1Kb), similarity, and orientation, are highly susceptible to duplications and deletions. These genomic rearrangements are caused by the mechanism of non-allelic homologous recombination. The resulting genomic variation leads to gene dosage dependent neurological disorders such as Rett-like syndrome and Pelizaeus-Merzbacher disease.
Gene duplication as amplification
Gene duplication doesn't necessarily constitute a lasting change in a species' genome. In fact, such changes often don't last past the initial host organism. From the perspective of molecular genetics, amplification is one of many ways in which a gene can be overexpressed. Genetic amplification can occur artificially, as with the use of the polymerase chain reaction technique to amplify short strands of DNA in vitro using enzymes, or it can occur naturally, as described above. If it's a natural duplication, it can still take place in a somatic cell, rather than a germline cell (which would be necessary for a lasting evolutionary change).
Also, in either event, duplications can be and often are marginally or severely detrimental. For instance, duplications of oncogenes are a common cause of many types of cancer, as with P70-S6 Kinase 1 amplification and breast cancer. In such cases the genetic duplication occurs in a somatic cell and affects only the genome of the cancer cells themselves, not the entire organism, much less any subsequent offspring.
Gene duplication
Gene duplication (or chromosomal duplication or gene amplification) is any duplication of a region of DNA that contains a gene; it may occur as an error in homologous recombination, a retrotransposition event, or duplication of an entire chromosome. The second copy of the gene is often free from selective pressure — that is, mutations of it have no deleterious effects to its host organism. Thus it mutates faster than a functional single-copy gene, over generations of organisms.
Gene duplication as an evolutionary event
Gene duplication is believed to play a major role in evolution; this stance has been held by members of the scientific community for over 100 years. Susumu Ohno was one of the most famous developers of this theory in his classic book Evolution by gene duplication (1970). Ohno argued that gene duplication is the most important evolutionary force since the emergence of the universal common ancestor. Major genome duplication events are not uncommon. It is believed that the entire yeast genome underwent duplication about 100 million years ago. Plants are the most prolific genome duplicators. For example, wheat is hexaploid (a kind of polyploid), meaning that it has six copies of its genome.
The two genes that exist after a gene duplication event are called paralogs and usually code for proteins with a different function and/or structure. By contrast, orthologous genes are ones which code for proteins with similar functions but exist in different species, and are created from a speciation event. (See Homology of sequences in genetics).
It is important (but often difficult) to differentiate between paralogs and orthologs in biological research. Experiments on human gene function can often be carried out on other species if a homolog to a human gene can be found in the genome of that species, but only if the homolog is orthologous. If they are paralogs and resulted from a gene duplication event, their functions are likely to be too different.
The paralogous segments can be repeat sequences with more than 90% sequence similarity. In such cases, they are known as low copy repeats (LCRs) though they are not highly repetitive sequences. They are mostly found in pericentronomic, subtelomeric and interstitial regions of a chromosome. The LCRs, due to their size (>1Kb), similarity, and orientation, are highly susceptible to duplications and deletions. These genomic rearrangements are caused by the mechanism of non-allelic homologous recombination. The resulting genomic variation leads to gene dosage dependent neurological disorders such as Rett-like syndrome and Pelizaeus-Merzbacher disease.
Gene duplication as amplification
Gene duplication doesn't necessarily constitute a lasting change in a species' genome. In fact, such changes often don't last past the initial host organism. From the perspective of molecular genetics, amplification is one of many ways in which a gene can be overexpressed. Genetic amplification can occur artificially, as with the use of the polymerase chain reaction technique to amplify short strands of DNA in vitro using enzymes, or it can occur naturally, as described above. If it's a natural duplication, it can still take place in a somatic cell, rather than a germline cell (which would be necessary for a lasting evolutionary change).
Also, in either event, duplications can be and often are marginally or severely detrimental. For instance, duplications of oncogenes are a common cause of many types of cancer, as with P70-S6 Kinase 1 amplification and breast cancer. In such cases the genetic duplication occurs in a somatic cell and affects only the genome of the cancer cells themselves, not the entire organism, much less any subsequent offspring.











No comments:
Post a Comment